Contents
Navigation:
Interaction energy calculations
Here we will learn how to perform simple interaction energy calculations with CamCASP.
Water dimer : SAPT(DFT) energy scan
Water dimer energy scan in its global minimum energy configuration. In this part of the tutorial we will perform a sequence of SAPT(DFT) interaction energy evaluations for the water dimer using a pre-defined set of dimer orientations.  In this case, we will place the two water molecules in their (near) global-minimum energy configuration and perform a scan along the O-O separation vector. This configuration is illustrated in the adjoining figure.
In this case, we will place the two water molecules in their (near) global-minimum energy configuration and perform a scan along the O-O separation vector. This configuration is illustrated in the adjoining figure.
Files needed
The SAPT(DFT) calculation is performed in two steps:
- Second-order interaction calculation using SAPT(DFT)
$\delta^{\rm HF}_{\rm int}$ calculation. This is needed for the (mainly) induction energy contributions from third- to infinite-order in the perturbation expansion.
We will use separate Cluster input files for each of these steps. And we will also need the geometry parameters provided in a separate file. These are displayed in this section. Please download the files by clicking on the file name headers.
Cluster file for SAPT(DFT) calculation:
Title water dimer cluster file
Global
Units Bohr degrees
End
MOLECULE water1
Units Bohr
!
! Experimental IP.
IP 0.4638 a.u.
! AC-shift computed using e_HOMO(aTZ/PBE0) = -0.3327 a.u.
AC-Shift 0.1311 a.u.
!
! Vibrationally averaged geom.
O1 8.0 0.00000000 0.00000000 0.00000000
H1 1.0 -1.45365196 0.00000000 -1.12168732
H2 1.0 1.45365196 0.00000000 -1.12168732
END
MOLECULE water2
Units Bohr
!
! Experimental IP.
IP 0.4638 a.u.
! AC-shift computed using e_HOMO(aTZ/PBE0) = -0.3327 a.u.
AC-Shift 0.1311 a.u.
!
! Vibrationally averaged geom.
O1 8.0 0.00000000 0.00000000 0.00000000
H1 1.0 -1.45365196 0.00000000 -1.12168732
H2 1.0 1.45365196 0.00000000 -1.12168732
END
Rotate water2 by {alpha} about {Nx} {Ny} {Nz}
Place water2 at {Rx} {Ry} {Rz}
Join water1 and water2 into water_dimer
Show water_dimer in XYZ format
Run-Type
SAPT(DFT)
Molecules water1 and water2
File-Prefix {job}
SCFcode NWChem
Method DFT
Func PBE0
AC CS00
Kernel ALDA+CHF
Basis aug-cc-pVTZ Type MC+
Aux-Basis aug-cc-pVTZ Type DC+
Mid-Bond 3s2p1d1f Type Weighted
Memory 2 GB
End
FinishIn this calculation we will perform a SAPT(DFT) interaction energy calculation using:
The PBE0 functional with the CS00 (Casida-Salahub 2000) asymptotic correction.
- The NWChem code will be used for the DFT calculation.
- The linear-response kernel will be ALDA+CHF (this is calculated within CamCASP).
- The main basis is the aug-cc-pVTZ basis in the MC+ format with a 3s2p1d1f mid-bond set.
- The auxiliary basis used in the density-fitting step will the RI-MP2-aug-cc-pVTZ basis in the DC+ form.
Important
To use Psi4 instead of NWChem use the following commands in the last part of the file:
Here we have replaced the **SCFcode** by **Psi4** and the **AC** (asymptotic correction) by **GRAC**.
Run-Type
SAPT(DFT)
Molecules water1 and water2
File-Prefix {job}
SCFcode Psi4
Method DFT
Func PBE0
AC GRAC
Kernel ALDA+CHF
Basis aug-cc-pVTZ Type MC+
Aux-Basis aug-cc-pVTZ Type DC+
Mid-Bond 3s2p1d1f Type Weighted
Memory 2 GB
EndNote
Variables in the Cluster file:
Notice that the geometry parameters for the rotation and translation of water2 have been specified within "{..}". These variables - using exactly the names shown above - will be replaced by the Python codes using values read in from the geometry file (shown below). Likewise, the FILE-PREFIX must be specified as {job} as it too will be replaced with a value.
Cluster file for Delta-HF energy calculation:
Title water dimer cluster file : delta-HF
Global
Units Bohr degrees
End
MOLECULE water1
Units Bohr
IP 0.4638 a.u.
! Vibrationally averaged geom.
O1 8.0 0.00000000 0.00000000 0.00000000
H1 1.0 -1.45365196 0.00000000 -1.12168732
H2 1.0 1.45365196 0.00000000 -1.12168732
END
MOLECULE water2
Units Bohr
IP 0.4638 a.u.
! Vibrationally averaged geom.
O1 8.0 0.00000000 0.00000000 0.00000000
H1 1.0 -1.45365196 0.00000000 -1.12168732
H2 1.0 1.45365196 0.00000000 -1.12168732
END
Rotate water2 by {alpha} about {Nx} {Ny} {Nz}
Place water2 at {Rx} {Ry} {Rz}
Join water1 and water2 into water_dimer
Show water_dimer in XYZ format
Run-Type
Delta-HF
Molecules water1 and water2
File-Prefix {job}
SCFcode NWChem
Basis aug-cc-pVTZ Type DC
Aux-Basis aug-cc-pVTZ Type DC
Memory 2 GB
End
FinishThis file is very similar to the one used for the SAPT(DFT) calculation, except for the following:
- This is a DELTA-HF calculation
- The basis sets are both of type DC. (MC+ or MC cannot be used here, and DC+ is superfluous as this energy correction is not sensitive to the mid-bond set)
- The DFT options have all been removed. Had they been present, they would have been ignored.
Important
To use Psi4 instead of NWChem use the following commands in the last part of the file:
Here we have replaced the **SCFcode** by **Psi4**.
Run-Type
Delta-HF
Molecules water1 and water2
File-Prefix {job}
SCFcode Psi4
Basis aug-cc-pVTZ Type DC
Aux-Basis aug-cc-pVTZ Type DC
Memory 2 GB
EndDimer geometry file for energy scan:
! UNITS BOHR ! UNITS DEGREES ! INDEX Rx Ry Rz alpha Nx Ny Nz 1 -4.03818248 0.0 -2.45198575 133.12505 -0.637206 0.637207 -0.433515 2 -4.36123708 0.0 -2.64814461 133.12505 -0.637206 0.637207 -0.433515 3 -4.52276438 0.0 -2.74622404 133.12505 -0.637206 0.637207 -0.433515 4 -4.68429168 0.0 -2.84430347 133.12505 -0.637206 0.637207 -0.433515 5 -4.83352211 0.0 -2.93491622 133.12505 -0.637206 0.637207 -0.433515 6 -5.00734628 0.0 -3.04046233 133.12505 -0.637206 0.637207 -0.433515 7 -5.16887358 0.0 -3.13854176 133.12505 -0.637206 0.637207 -0.433515 8 -5.33040088 0.0 -3.23662119 133.12505 -0.637206 0.637207 -0.433515 9 -5.65345548 0.0 -3.43278005 133.12505 -0.637206 0.637207 -0.433515 10 -6.46109198 0.0 -3.92317720 133.12505 -0.637206 0.637207 -0.433515 11 -8.07636497 0.0 -4.90397150 133.12505 -0.637206 0.637207 -0.433515 12 -9.69163797 0.0 -5.88476580 133.12505 -0.637206 0.637207 -0.433515
Here the geometry is specified as indicated in the line with the column headers. The units should be identical to those set in the GLOBAL..END part of the Cluster command files: in this case, BOHR and DEGREES.
Commands to run the jobs
The CamCASP suite of codes comes with a set of useful Python codes to help perform various calculations. The one we will use here is batch_camcasp.py. Check to see if you have access to it by doing the following:
$ batch_camcasp.py --help
usage: batch_camcasp.py [-h] [--dHF] [-b BASIS] [-t {mc,mc+,dc,dc+}]
[--direct] [--memory MEMORY] [-q {bg,batch,none}]
[--scfcode {dalton2006,dalton,dalton2013,nwchem,}]
[--nproc NPROC] [--core CORES]
job template geomfile
Carry out a batch of SAPT-DFT or delta-HF calculations.
positional arguments:
job job name
template cluster file template
geomfile file containing geometry data
optional arguments:
-h, --help show this help message and exit
--dHF run delta-HF calculations instead of sapt-dft
-b BASIS, --basis BASIS
basis set for calculation (default aug-cc-pVTZ)
-t {mc,mc+,dc,dc+}, --type {mc,mc+,dc,dc+}
basis type (default mc+)
--direct Use direct integral management
--memory MEMORY, -M MEMORY
Specify memory for job in MB
-q {bg,batch,none}, --queue {bg,batch,none}
Specify queue for job (default batch)
--scfcode {dalton2006,dalton,dalton2013,nwchem,}
Force use of scfcode
--nproc NPROC Number of processors to use
--core CORES Number of cores on machine
More text follows...The actual help is longer than this, but I have included the main options above. This code will extract the geometry parameters from the geometry file and insert them in the Cluster file and run the runcamcasp.py code that will actually do the work of setting up the individual jobs.
Important
If you do not get the above output and instead get a command-not-found error, then your PATH variable is probably not set up to give you access to this - and presumably other - codes from the CamCASP installation.
If you get the above output, then proceed to the next step.
First check to see that all is well using. This is important as it is easy to make mistakes in the input files. We do this by using the option -q none. This tells the codes to try to set up all input files, but not to run them. Thereby giving you a chance to see it they were setup correctly.
batch_camcasp.py -q none water2 water2_saptdft_scan.clt globalminconfig_ordered.geom
Here's what you should find in your directory:
$ ls globalminconfig_ordered.geom water2_11 water2_1.clt water2_3.clt water2_5.clt water2_7.clt water2_9.clt water2_1 water2_11.clt water2_2 water2_4 water2_6 water2_8 water2_saptdft_scan.clt water2_10 water2_12 water2_2.clt water2_4.clt water2_6.clt water2_8.clt water2_10.clt water2_12.clt water2_3 water2_5 water2_7 water2_9
Here's what should be in one of the sub-directories:
$ cd water2_1 $ ls submit.py water2_1_B.nw water2_1.clt water2_1_energy.ornt water_dimer.pdb water2_1_A.nw water2_1.cks water2_1.clt.clout water2_1.sh
If the files look OK (no obvious errors - you do need to look into them to make sure that they correspond to the kind of calculation you were interested in), then you can run them by either going into each sub-directory and running the submit.py script, like so:
$ cd water2_1 $ ./submit.py -q batch
Here we have submitted the job to the batch queue. You will need to do this for each of the directories. This can be tedious, but may be a useful way to run a single job to make sure all is well.
Important
Some comments on the batch queue:
- This queue will run jobs only if the load on the system is less than a pre-set value. This is normally set to be slightly less than the number of processors on the system.
- Useful commands: atq, atrm
man batch will give you a full list of commands.
Alternatively, if you do not fancy going into each directory and typing ./submit.py -q batch, do the following:
$ ls
globalminconfig_ordered.geom water2_11 water2_1.clt water2_3.clt water2_5.clt water2_7.clt water2_9.clt
water2_1 water2_11.clt water2_2 water2_4 water2_6 water2_8 water2_saptdft_scan.clt
water2_10 water2_12 water2_2.clt water2_4.clt water2_6.clt water2_8.clt
water2_10.clt water2_12.clt water2_3 water2_5 water2_7 water2_9
$ rm -Rf water2_{1..12}*
$ ls
globalminconfig_ordered.geom water2_saptdft_scan.clt
$ batch_camcasp.py -q batch water2 water2_saptdft_scan.clt globalminconfig_ordered.geomImportant
Here we have removed all the directories created by the codes before running the batch_camcasp.py code, this time with -q batch. The remove step is needed as batch_camcasp.py is conservative: if it finds directories with names identical to the ones it will create, it will not proceed and instead will give you an error like this:
$ batch_camcasp.py -q batch water2 water2_saptdft_scan.clt globalminconfig_ordered.geom ['runcamcasp.py', 'water2_1', '-d', 'water2_1', '--ifexists', 'abort', '-q', 'batch', '--direct', '--scfcode', 'nwchem', '--nproc', '2'] Directory water2_1 exists; aborting job
Important
If the jobs are running correctly you will see them listed if you run
- $ top
or
- $ htop
If they are not running, then something is wrong and you should check all error/warning and *.out files in the working directories.
If they have run successfully, you will see the following in each working directory:
- $ cd water2_1 $ ls OUT water2_1_A.nw water2_1_B.nw water2_1.clt water2_1_energy.ornt water_dimer.pdb water2_1-A-asc.movecs water2_1-B-asc.movecs water2_1.cks water2_1.clt.clout water2_1.sh $ cd OUT $ ls warnings.log water2_1_B.out water2_1.clt water2_1_energy.ornt water2_1.out water_dimer.pdb water2_1_A.out water2_1.cks water2_1-data-summary.data water2_1.log water2_1.sh
The NWChem DFT outputs will be in files water2_1_A.out and water2_1_B.out, and the CamCASP SAPT(DFT) output in file water2_1.out.
Once you have got the SAPT(DFT) calculations going, start the Delta-HF calculations using:
Running the Delta-HF calculations
$ batch_camcasp.py --dHF -q batch water2 water2_deltahf_scan.clt globalminconfig_ordered.geom
Now you should have the following files/directories present:
$ ls globalminconfig_ordered.geom water2_11.clt water2_1_dHF water2_3_dHF water2_5_dHF water2_7_dHF water2_9_dHF water2_1 water2_11_dHF water2_2 water2_4 water2_6 water2_8 water2_deltahf_scan.clt water2_10 water2_12 water2_2.clt water2_4.clt water2_6.clt water2_8.clt water2_saptdft_scan.clt water2_10.clt water2_12.clt water2_2_dHF water2_4_dHF water2_6_dHF water2_8_dHF water2_10_dHF water2_12_dHF water2_3 water2_5 water2_7 water2_9 water2_11 water2_1.clt water2_3.clt water2_5.clt water2_7.clt water2_9.clt
Extracting energies from the output files
The output files contain all the data you need and you can, in principle view these files and extract the SAPT(DFT) interaction energies for yourself. For example, have a look at the CamCASP output files for job water2_1. A summary of the results should be right at the end of the file and should look like this:
$ cd water2_1/OUT/
$ tail -n 30 water2_1.out
==
Name Value Units Description Misc(optional)
=====
E^{1}_{elst} -0.6677896E+04 CM-1 mol-mol :: DF :: NoReg
E^{1}_{exch}(S2) 0.1127260E+05 CM-1 DF :: S2 :: NoReg ::
E^{1}_{exch} 0.1157911E+05 CM-1 DF :: NoReg ::
E^{1}_{int} 0.4901212E+04 CM-1 DF :: NoReg ::
E^{2}_{ind} -0.5676361E+04 CM-1 DF :: PROP cks :: NoReg
E^{2}_{ind}(A) -0.2491498E+04 CM-1 DF :: PROP cks :: NoReg
E^{2}_{ind}(B) -0.3184863E+04 CM-1 DF :: PROP cks :: NoReg
E^{2}_{ind}(UC) -0.5412026E+04 CM-1 DF :: PROP UC :: NoReg
E^{2}_{ind,exch} 0.3946972E+04 CM-1 DF :: AMP C :: NoReg :: PROP cks
E^{2}_{ind,exch}(A) 0.2238703E+04 CM-1 DF :: AMP C :: NoReg :: PROP cks
E^{2}_{ind,exch}(B) 0.1708268E+04 CM-1 DF :: AMP C :: NoReg :: PROP cks
E^{2}_{ind,exch}(UC) 0.0000000E+00 CM-1 DF :: AMP UC :: :: NoReg
E^{2}_{disp} -0.2579572E+04 CM-1 DF :: NoReg :: PROP cks
E^{2}_{disp}(UC) -0.3361664E+04 CM-1 DF :: NoReg :: PROP UC
E^{2}_{disp,exch} 0.6294896E+03 CM-1 DF :: NoReg :: PROP scaled cks
E^{2}_{disp,exch}(UC) 0.8203427E+03 CM-1 DF :: NoReg :: PROP UC
E^{2}_{ind} -0.1095541E+04 CM-1 DF :: PROP cks :: REG eta = 3.0000
E^{2}_{ind}(A) 0.1495754E+02 CM-1 DF :: PROP cks :: REG eta = 3.0000
E^{2}_{ind}(B) -0.1110498E+04 CM-1 DF :: PROP cks :: REG eta = 3.0000
E^{2}_{ind}(UC) -0.1149311E+04 CM-1 DF :: PROP UC :: REG eta = 3.0000
E^{2}_{ind,exch} 0.1804795E+03 CM-1 DF :: AMP C :: REG eta = 3.0000 :: PROP cks
E^{2}_{ind,exch}(A) -0.1211432E+03 CM-1 DF :: AMP C :: REG eta = 3.0000 :: PROP cks
E^{2}_{ind,exch}(B) 0.3016227E+03 CM-1 DF :: AMP C :: REG eta = 3.0000 :: PROP cks
E^{2}_{ind,exch}(UC) 0.0000000E+00 CM-1 DF :: AMP UC :: :: REG eta = 3.0000
--------------------------------------------------------------------------------
Ending on 09-03-2016 at 18:16:46.186There you have it: all the energy components, energies, units, and a descriptor string. The SAPT(DFT) second-order interaction energy calculation is defined as: $ E{(2)}_{\rm int} = E{(1)}_{\rm elst} + E{(1)}_{\rm exch} + E{(2)}_{\rm ind} + E{(2)}_{\rm ind,exch} + E{(2)}_{\rm disp} + E^{(2)}_{\rm disp,exch} $
The $\delta^{\rm HF}_{\rm int}$ energy isn't included here. This energy must be assembled from calculations in water2_1_dHF/. This time you need not only the SAPT energies (not SAPT(DFT)!), but also the HF energies for the two monomers and the dimer:
$ cd water2_1_dHF/OUT/
$ tail -20 water2_1.out
Summary of CamCASP calculation
==
Name Value Units Description Misc(optional)
=====
E^{1}_{elst} -0.6732864E+04 CM-1 mol-mol :: DF :: NoReg
E^{1}_{exch}(S2) 0.1023535E+05 CM-1 DF :: S2 :: NoReg ::
E^{1}_{exch} 0.1049827E+05 CM-1 DF :: NoReg ::
E^{1}_{int} 0.3765406E+04 CM-1 DF :: NoReg ::
E^{2}_{ind} -0.4776384E+04 CM-1 DF :: PROP chf :: NoReg
E^{2}_{ind}(A) -0.1939896E+04 CM-1 DF :: PROP chf :: NoReg
E^{2}_{ind}(B) -0.2836489E+04 CM-1 DF :: PROP chf :: NoReg
E^{2}_{ind}(UC) -0.3931261E+04 CM-1 DF :: PROP UC :: NoReg
E^{2}_{ind,exch} 0.3058315E+04 CM-1 DF :: AMP C :: NoReg :: PROP chf
E^{2}_{ind,exch}(A) 0.1669277E+04 CM-1 DF :: AMP C :: NoReg :: PROP chf
E^{2}_{ind,exch}(B) 0.1389038E+04 CM-1 DF :: AMP C :: NoReg :: PROP chf
E^{2}_{ind,exch}(UC) 0.0000000E+00 CM-1 DF :: AMP UC :: :: NoReg
--------------------------------------------------------------------------------
Ending on 09-03-2016 at 18:31:24.375
$ grep 'Total SCF energy' water2_1_*.out
water2_1_AB.out: Total SCF energy = -152.115579524085
water2_1_A.out: Total SCF energy = -76.059372032942
water2_1_B.out: Total SCF energy = -76.059417963167Now define the following:
Define the HF interaction energy: $E^{\rm HF}_{\rm int} = E[AB] - E[A] - E[B]$
Convert $E^{\rm HF}_{\rm int}$ to cm$^{-1}$ to be consistent with the units used in the CamCASP output.
Define the corresponding second-order HF energy from the SAPT components: $E^{(2)}_{\rm HF,int} = E^{(1)}_{\rm elst} + E^{(1)}_{\rm exch} + E^{(2)}_{\rm ind} + E^{(2)}_{\rm ind,exch}$.
Define the DELTA-HF correction: $\delta^{\rm HF}_{\rm int} = E^{\rm HF}_{\rm int} - E^{(2)}_{\rm HF,int}$.
It is nice to know how to do this, but performing these calculations for all dimer configurations is tedious and is bound to be error-prone. So we have a Python code to do the work for us. This one is called extract_saptdft.py and has the following help page:
$ extract_saptdft.py --help
usage: extract_saptdft.py [-h] [--verbose]
[--unit {cm-1,kJ/mol,au,hartree,eV,meV,K,kelvin}]
[--S2 | --short | --long] [--ab] [--suffix SUFFIX]
[--dhf DHF]
path [path ...]
Extract sapt-dft energy terms from CamCASP summary files.
positional arguments:
path path to job directory (may be repeated)
optional arguments:
-h, --help show this help message and exit
--verbose, -v verbose output
--unit {cm-1,kJ/mol,au,hartree,eV,meV,K,kelvin}, --units {cm-1,kJ/mol,au,hartree,eV,meV,K,kelvin}
Unit for output (default kJ/mol)
--S2 Use S2 approximation for E(1)exch
--short Short output layout
--long Long output layout
--ab Show A and B induction separately
--suffix SUFFIX summary file suffix (default -data-summary.data)
--dhf DHF default delta-HF energy
More text follows...We will use it as follows:
$ ls globalminconfig_ordered.geom water2_11.clt water2_1_dHF water2_3_dHF water2_5_dHF water2_7_dHF water2_9_dHF water2_1 water2_11_dHF water2_2 water2_4 water2_6 water2_8 water2_deltahf_scan.clt water2_10 water2_12 water2_2.clt water2_4.clt water2_6.clt water2_8.clt water2_saptdft_scan.clt water2_10.clt water2_12.clt water2_2_dHF water2_4_dHF water2_6_dHF water2_8_dHF water2_10_dHF water2_12_dHF water2_3 water2_5 water2_7 water2_9 water2_11 water2_1.clt water2_3.clt water2_5.clt water2_7.clt water2_9.clt $ $ extract_saptdft.py water2_* kJ/mol elst exch ind exind dHF disp exdisp total water2_1 -79.88525 138.51670 -67.90425 47.21620 -16.06246 -30.85849 7.53036 -1.44719 water2_10 -7.49606 0.72324 -0.70207 0.12027 0.05605 -1.35946 0.07266 -8.58537 water2_11 -3.23829 0.02026 -0.18011 0.00285 0.04701 -0.26896 0.00266 -3.61458 water2_12 -1.70827 0.00057 -0.05517 0.00007 0.02144 -0.07633 0.00010 -1.81760 water2_2 -51.26629 69.87602 -32.14671 21.32642 -7.94215 -19.72405 4.31327 -15.56349 water2_3 -41.66366 49.51204 -22.46926 14.28243 -5.49456 -15.80083 3.22541 -18.40841 water2_4 -34.22959 35.03832 -15.85054 9.54977 -3.79562 -12.67985 2.39703 -19.57049 water2_5 -28.83334 25.42702 -11.56678 6.57399 -2.69150 -10.36566 1.81364 -19.64263 water2_6 -23.90075 17.47970 -8.10563 4.25782 -1.78447 -8.21776 1.30445 -18.96665 water2_7 -20.30716 12.32561 -5.89850 2.84873 -1.20097 -6.64016 0.95647 -17.91598 water2_8 -17.43745 8.68310 -4.35104 1.90798 -0.79830 -5.38078 0.69897 -16.67752 water2_9 -13.22459 4.29450 -2.44350 0.85858 -0.31987 -3.56681 0.36991 -14.03178
That's it. All done. The default energy units are 'kJ/mol' - which is far more sensible than cm$^{-1}$.
Important
To send the output of the extract code to another file use:
It is always sensible to use a file name that tells you what is in it. }}}
You may want to do is to include some or all of the geometry parameters in this file. Here is what my own energy file looks like:
#kJ/mol Rx Ry Rz elst exch ind exind dHF disp exdisp total 1 -4.03818248 0.0 -2.45198575 -79.88525 138.51670 -67.90425 47.21620 -16.06246 -30.85849 7.53036 -1.44719 2 -4.36123708 0.0 -2.64814461 -51.26629 69.87602 -32.14671 21.32642 -7.94215 -19.72405 4.31327 -15.56349 3 -4.52276438 0.0 -2.74622404 -41.66366 49.51204 -22.46926 14.28243 -5.49456 -15.80083 3.22541 -18.40841 4 -4.68429168 0.0 -2.84430347 -34.22959 35.03832 -15.85054 9.54977 -3.79562 -12.67985 2.39703 -19.57049 5 -4.83352211 0.0 -2.93491622 -28.83334 25.42702 -11.56678 6.57399 -2.69150 -10.36566 1.81364 -19.64263 6 -5.00734628 0.0 -3.04046233 -23.90075 17.47970 -8.10563 4.25782 -1.78447 -8.21776 1.30445 -18.96665 7 -5.16887358 0.0 -3.13854176 -20.30716 12.32561 -5.89850 2.84873 -1.20097 -6.64016 0.95647 -17.91598 8 -5.33040088 0.0 -3.23662119 -17.43745 8.68310 -4.35104 1.90798 -0.79830 -5.38078 0.69897 -16.67752 9 -5.65345548 0.0 -3.43278005 -13.22459 4.29450 -2.44350 0.85858 -0.31987 -3.56681 0.36991 -14.03178 10 -6.46109198 0.0 -3.92317720 -7.49606 0.72324 -0.70207 0.12027 0.05605 -1.35946 0.07266 -8.58537 11 -8.07636497 0.0 -4.90397150 -3.23829 0.02026 -0.18011 0.00285 0.04701 -0.26896 0.00266 -3.61458 12 -9.69163797 0.0 -5.88476580 -1.70827 0.00057 -0.05517 0.00007 0.02144 -0.07633 0.00010 -1.81760
Plotting the energies is quite easy once you have the data in a file such as this.
Important
In these calculations we have used SAPT(DFT) based on PBE0 with the CS00 (Casida-Salahub) asymptotic correction with the default shift values. This is not ideal for a few reasons:
- - The actual shift should be used; not the default value.
- The CS00 and GRAC (both based on LB94) asymptotic corrections may not be as suitable for small dimers as the Fermi-Amaldi correction with the Tozer-Handy splicing. Tests done a long time ago (see papers by Misquitta & Szalewicz in 2005) suggested that the XC-potential from LB94 functional was a bit too repulsive at intermediate range even though it did tend to $-1/r$ at long-range.
This may help explain why the interaction energies are too small by about 1-1.5 kJ/mol. Of course, some of this error is due to the basis set: the aTZ MC+ basis is a good one, but the aQZ MC+ basis would have been a better choice here.
Analysis
A sensible analysis of the data might involve making a plot of the energies versus COM..COM separation $R$. Rather than plot all energy components, some may be grouped together as:
$E^{(2)}_{\rm IND} = E^{(2)}_{\rm ind} + E^{(2)}_{\rm ind,exch}$
$E^{(2)}_{\rm DISP} = E^{(2)}_{\rm disp} + E^{(2)}_{\rm disp,exch}$
- We do not normally group the first-order energies together, though it may be argued that there is some reason to do so.
Here is a plot of the above data: 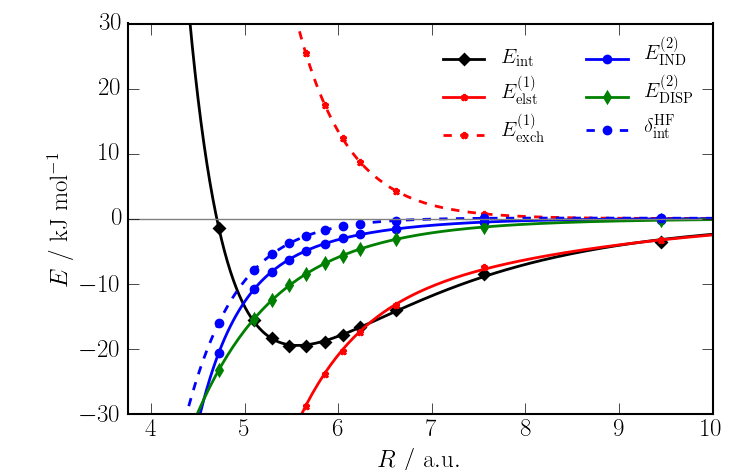 This very clearly shows that
This very clearly shows that
- The results make sense! The energies follow a pattern.
The interaction energy from the minimum onwards is dominated by $E^{(1)}_{\rm elst}$.
- The second-order dispersion dominates the second-order induction.
But the total induction energy - which includes the $\delta^{\rm HF}_{\rm int}$ energy as it is mostly induction - is comparable to the dispersion energy in importance in determining the binding of this system.
So far there is no mention of charge-transfer: SAPT(DFT) does not separate out the charge-transfer energy from the polarization term; instead, both are included in the induction energy. To perform this separation we use Reg-SAPT(DFT) as described on another page on this Wiki.
The pyridine dimers
Data for some dimers found on Model(3) [ref needed].
Cluster file for the SAPT(DFT) calculation:
Title Pyridine : SAPT(DFT) scan file
Global
Units Bohr Degree
Overwrite Yes
End
Molecule pyr1
! Optimzed with PBE0/cc-pVTZ Gaussian03
! C2v symmetry
IP 0.3488 a.u.
Units Angstrom
H1 1.0 -2.050322 1.274414 0.000000
H2 1.0 -2.147113 -1.203259 0.000000
H3 1.0 0.000000 -2.487558 0.000000
H4 1.0 2.147113 -1.203259 0.000000
H5 1.0 2.050322 1.274414 0.000000
N 7.0 0.000000 1.382844 0.000000
C1 6.0 -1.134410 0.690452 0.000000
C2 6.0 -1.190513 -0.695795 0.000000
C3 6.0 0.000000 -1.403912 0.000000
C4 6.0 1.190513 -0.695795 0.000000
C5 6.0 1.134410 0.690452 0.000000
End
Molecule pyr2
! Optimzed with PBE0/cc-pVTZ Gaussian03
! C2v symmetry
IP 0.3488 a.u.
Units Angstrom
H1 1.0 -2.050322 1.274414 0.000000
H2 1.0 -2.147113 -1.203259 0.000000
H3 1.0 0.000000 -2.487558 0.000000
H4 1.0 2.147113 -1.203259 0.000000
H5 1.0 2.050322 1.274414 0.000000
N 7.0 0.000000 1.382844 0.000000
C1 6.0 -1.134410 0.690452 0.000000
C2 6.0 -1.190513 -0.695795 0.000000
C3 6.0 0.000000 -1.403912 0.000000
C4 6.0 1.190513 -0.695795 0.000000
C5 6.0 1.134410 0.690452 0.000000
End
Rotate pyr2 by {alpha} about {Nx} {Ny} {Nz}
Place pyr2 at {Rx} {Ry} {Rz}
Files
SAPT(DFT)
Molecule pyr1 and pyr2
Basis augA-Sadlej Type MC+
MidBond 3s3p2d2f Type Weighted
Aux-Basis aTZ Type DC+
DFTcode DALTON2006
AC MULTPOLE
Func PBE0
Kernel ALDA+CHF
File-prefix {job}
Interface file
Memory 6 GB
Options NO-3rd-Order
End
FinishCluster file for the $\delta^{\rm HF}_{\rm int}$ calculation:
Title Pyridine : deltaHF scan file
Global
Units Bohr Degree
Overwrite Yes
End
Molecule pyr1
! Optimzed with PBE0/cc-pVTZ Gaussian03
! C2v symmetry
IP 0.3488 a.u.
Units Angstrom
H1 1.0 -2.050322 1.274414 0.000000
H2 1.0 -2.147113 -1.203259 0.000000
H3 1.0 0.000000 -2.487558 0.000000
H4 1.0 2.147113 -1.203259 0.000000
H5 1.0 2.050322 1.274414 0.000000
N 7.0 0.000000 1.382844 0.000000
C1 6.0 -1.134410 0.690452 0.000000
C2 6.0 -1.190513 -0.695795 0.000000
C3 6.0 0.000000 -1.403912 0.000000
C4 6.0 1.190513 -0.695795 0.000000
C5 6.0 1.134410 0.690452 0.000000
End
Molecule pyr2
! Optimzed with PBE0/cc-pVTZ Gaussian03
! C2v symmetry
IP 0.3488 a.u.
Units Angstrom
H1 1.0 -2.050322 1.274414 0.000000
H2 1.0 -2.147113 -1.203259 0.000000
H3 1.0 0.000000 -2.487558 0.000000
H4 1.0 2.147113 -1.203259 0.000000
H5 1.0 2.050322 1.274414 0.000000
N 7.0 0.000000 1.382844 0.000000
C1 6.0 -1.134410 0.690452 0.000000
C2 6.0 -1.190513 -0.695795 0.000000
C3 6.0 0.000000 -1.403912 0.000000
C4 6.0 1.190513 -0.695795 0.000000
C5 6.0 1.134410 0.690452 0.000000
End
Rotate pyr2 by {alpha} about {Nx} {Ny} {Nz}
Place pyr2 at {Rx} {Ry} {Rz}
Files
DeltaHF
Molecule pyr1 and pyr2
Basis augA-Sadlej Type DC
Aux-Basis aTZ Type DC
DFTcode DALTON2006
Kernel CHF
File-prefix {job}
Interface file
Memory 6 GB
Options NO-3rd-Order
End
Finish
Useful scripts for running the jobs
Make sure these are executable. You do this using:
$ chmod +x <script name>
Now you should be able to execute it using:
$ ./<script name> <name of geometry file>
This one is to run a SAPT(DFT) job. It uses the file //pyridine2-saptdft-scan.clt// and one of the geometry files given below:
geom=$1 if [[ ! -a $geom ]]; then echo "No geometry file specified. geom = $geom" exit -1 fi batch_camcasp.py --direct --nproc 2 --scfcode dalton2006 -q batch pyr2 pyridine2-saptdft-scan.clt $geom
This one is for a $\delta^{\rm HF}_{\rm int}$ calculation:
geom=$1 if [[ ! -a $geom ]]; then echo "No geometry file specified. geom = $geom" exit -1 fi batch_camcasp.py --direct --nproc 3 --scfcode dalton2006 -q batch --dHF pyr2 pyridine2-deltahf-scan.clt $geom
Here are geometry files for four of the pyridine dimers.
Min 1 : Hb1
! CGF = 13
! Config List name : CfgList-1
! Configs read from file : run1/pyr2_L2_C10iso_CT_Relax12_n2.geom
! UNITS BOHR DEGREE KJ/MOL
1 4.65059310 7.40505353 -0.00004327 180.00072878 -1.00000000 0.00000201 0.00000200
2 4.94125516 7.86786937 -0.00004598 180.00072878 -1.00000000 0.00000201 0.00000200
3 5.23191723 8.33068522 -0.00004868 180.00072878 -1.00000000 0.00000201 0.00000200
4 5.52257930 8.79350106 -0.00005139 180.00072878 -1.00000000 0.00000201 0.00000200
5 5.81324137 9.25631691 -0.00005409 180.00072878 -1.00000000 0.00000201 0.00000200
6 6.10390344 9.71913275 -0.00005680 180.00072878 -1.00000000 0.00000201 0.00000200
7 6.39456551 10.18194860 -0.00005950 180.00072878 -1.00000000 0.00000201 0.00000200
8 6.97588964 11.10758029 -0.00006491 180.00072878 -1.00000000 0.00000201 0.00000200
9 8.13853792 12.95884367 -0.00007573 180.00072878 -1.00000000 0.00000201 0.00000200
Min 2 : S1
! CGF = 6
! Config List name : CfgList-1
! Configs read from file : run1/pyr2_L2_C10iso_CT_Relax12_n2.geom
! UNITS BOHR DEGREE KJ/MOL
1 -1.84581291 -0.78492199 5.13854585 180.00061331 0.00000009 0.00000113 -1.00000000
2 -1.96117622 -0.83397962 5.45970497 180.00061331 0.00000009 0.00000113 -1.00000000
3 -2.07653953 -0.88303724 5.78086409 180.00061331 0.00000009 0.00000113 -1.00000000
4 -2.19190283 -0.93209487 6.10202320 180.00061331 0.00000009 0.00000113 -1.00000000
5 -2.30726614 -0.98115249 6.42318232 180.00061331 0.00000009 0.00000113 -1.00000000
6 -2.42262945 -1.03021012 6.74434143 180.00061331 0.00000009 0.00000113 -1.00000000
7 -2.53799275 -1.07926774 7.06550055 180.00061331 0.00000009 0.00000113 -1.00000000
8 -2.76871937 -1.17738299 7.70781878 180.00061331 0.00000009 0.00000113 -1.00000000
9 -3.23017260 -1.37361349 8.99245525 180.00061331 0.00000009 0.00000113 -1.00000000
Min 3 : S2
! CGF = 12
! Config List name : CfgList-1
! Configs read from file : run1/pyr2_L2_C10iso_CT_Relax12_n2.geom
! UNITS BOHR DEGREE KJ/MOL
1 1.11208672 -1.20581370 -5.18896550 179.99977143 0.81784921 0.57390414 0.04191305
2 1.18159214 -1.28117706 -5.51327585 179.99977143 0.81784921 0.57390414 0.04191305
3 1.25109756 -1.35654041 -5.83758619 179.99977143 0.81784921 0.57390414 0.04191305
4 1.32060298 -1.43190377 -6.16189653 179.99977143 0.81784921 0.57390414 0.04191305
5 1.39010840 -1.50726713 -6.48620688 179.99977143 0.81784921 0.57390414 0.04191305
6 1.45961382 -1.58263048 -6.81051722 179.99977143 0.81784921 0.57390414 0.04191305
7 1.52911924 -1.65799384 -7.13482756 179.99977143 0.81784921 0.57390414 0.04191305
8 1.66813009 -1.80872055 -7.78344825 179.99977143 0.81784921 0.57390414 0.04191305
9 1.94615177 -2.11017398 -9.08068963 179.99977143 0.81784921 0.57390414 0.04191305
Min 4 : T1
! CGF = 23
! Config List name : CfgList-1
! Configs read from file : run1/pyr2_L2_C10iso_CT_Relax12_n2.geom
! UNITS BOHR DEGREE KJ/MOL
1 -0.00107017 2.08464044 -6.95582290 176.48461160 -0.70688173 0.03057341 -0.70667071
2 -0.00113706 2.21493047 -7.39056183 176.48461160 -0.70688173 0.03057341 -0.70667071
3 -0.00120394 2.34522049 -7.82530076 176.48461160 -0.70688173 0.03057341 -0.70667071
4 -0.00127083 2.47551052 -8.26003969 176.48461160 -0.70688173 0.03057341 -0.70667071
5 -0.00133771 2.60580055 -8.69477862 176.48461160 -0.70688173 0.03057341 -0.70667071
6 -0.00140460 2.73609058 -9.12951756 176.48461160 -0.70688173 0.03057341 -0.70667071
7 -0.00147148 2.86638060 -9.56425649 176.48461160 -0.70688173 0.03057341 -0.70667071
8 -0.00160526 3.12696066 -10.43373435 176.48461160 -0.70688173 0.03057341 -0.70667071
9 -0.00187280 3.64812077 -12.17269007 176.48461160 -0.70688173 0.03057341 -0.70667071
Min 5 : T2
! CGF = 18
! Config List name : CfgList-1
! Configs read from file : run1/pyr2_L2_C10iso_CT_Relax12_n2.geom
! UNITS BOHR DEGREE KJ/MOL
1 -0.00118706 2.30100313 6.96310263 134.10426443 0.64045229 0.42340876 0.64073855
2 -0.00126125 2.44481583 7.39829655 134.10426443 0.64045229 0.42340876 0.64073855
3 -0.00133544 2.58862853 7.83349046 134.10426443 0.64045229 0.42340876 0.64073855
4 -0.00140963 2.73244122 8.26868438 134.10426443 0.64045229 0.42340876 0.64073855
5 -0.00148383 2.87625392 8.70387829 134.10426443 0.64045229 0.42340876 0.64073855
6 -0.00155802 3.02006661 9.13907220 134.10426443 0.64045229 0.42340876 0.64073855
7 -0.00163221 3.16387931 9.57426612 134.10426443 0.64045229 0.42340876 0.64073855
8 -0.00178059 3.45150470 10.44465395 134.10426443 0.64045229 0.42340876 0.64073855
9 -0.00207736 4.02675548 12.18542961 134.10426443 0.64045229 0.42340876 0.64073855